Simulating 500 million years of natural evolution, a new protein model ESM3 was born! Former Meta veteran LeCun likes this masterpiece

Following the update of AlphaFold 3, we saw another large model ESM3 in the field of life sciences.
The model development team comes from a startup called Evolutionary Scale AI. Team leader Alex Rives officially announced the release of the model on Twitter.

This exciting news was also forwarded by Yann LeCun, who said that your company is a bit like "making a fortune in silence."

What are the competitive advantages of ESM3 compared to the AlphaFold series?
The first is something the Meta team is familiar with – open source.

Although the model API is still in the beta stage and requires application for trial qualification, the model code has been put on GitHub. In addition, the company will cooperate with AWS and NVIDIA cloud computing platforms to facilitate developers to use and deploy.
Repository address: https://github.com/evolutionaryscale/esm
Unfortunately, the model weights are not yet public in the HuggingFace repository. Nvidia's official blog shows that ESM3 will provide a small open source version of the code and weights on the Nvidia BieNeMo platform, but only for non-commercial use.
Repository address: https://huggingface.co/EvolutionaryScale/esm3-sm-open-v1/tree/main
In addition, unlike AlphaFold 3, which simulates a variety of biological molecules, ESM3 only focuses on proteins, but can simultaneously infer their sequence, structure and function. This multimodal capability is a first in the field.
What is even more refreshing is that ESM3 was trained on 2.78 billion diverse proteins in nature, and gradually learned how the evolutionary process causes proteins to change.
From this perspective, the reasoning process of ESM can be regarded as an "evolution simulator", which opens up a new perspective for current life science research. The team even proposed the slogan "Simulating 500 million years of evolution" in the official website article.
Perhaps you have noticed that the name ESM is very similar to Meta's previous protein model ESMFold.
This is not an intentional borderline. In fact, the startup Evolutionary Scale was founded by former members of the Meta-FAIR protein group, and the company's chief scientist Alex Rives was the former head of the now-disbanded group.

Last August, in Meta’s “year of efficiency,” Zuckerberg chose to disband the protein group of only a dozen scientists to allow the company to focus on more profitable research.

But Rives was not intimidated by Meta's move and decided to start his own business. They have now raised $142 million in seed funding.
So let’s take a closer look at what new content this time in ESM3?

ESM3: Cutting-edge language models for biology
Life sciences are not as mysterious and unpredictable as we imagine.
Although protein molecules have incredible diversity and dynamic changes, their synthesis follows strict algorithms and processes. If it is regarded as a technology, its level of advancement far exceeds any engineering created by humans.
Biology is a thick code book.
It’s just that the codebook is written in a language we don’t yet understand, and even the tools running on today’s most powerful supercomputers can only scratch the surface.
If humans can read, or even write, the "code of life," biology can be made programmable. Trial and error will be replaced by logic, and laborious experiments will be replaced by simulations.
ESM3 is a step towards this grand vision and is the first generative model to date that can simultaneously reason about protein sequence, structure and function.
The rapid development of LLM in the past five years has also allowed the ESM team to discover the power of Scaling Law. They found that the same model also applies to biology.
As the size of training data and parameters increases, the model will gain a deeper understanding of basic biological principles and be able to better predict and design biological structures and functions.
Therefore, the development idea of ESM3 is also in line with the Scaling Law. Its scale is greatly expanded compared with the previous generation ESM. The amount of data has increased by 60 times, the amount of training calculation has increased by 25 times, and it is a generative model with native multimodality.
ESM3 was trained on the diversity of Earth's natural environments - billions of proteins, from the Amazon rainforest to the depths of the ocean, from microorganisms in the soil to extremes like deep-sea hydrothermal vents.
The model card on HuggingFace shows that the number of natural proteins in the training set reached 2.78 billion, and was enhanced to 3.15 billion sequences, 2.36 structures, and 539 million proteins with functional annotations through synthetic data, with a total token number of 7710B.
The total number of model training parameters reached 98B, using more than 10 24 FLOPS of computing power. The team seems to have worked closely with NVIDIA, and the Andromeda cluster was used for training, which is one of the GPU clusters with the highest throughput today, deploying the most advanced H100 GPU and Quantum-2 InfiniBand network.

Web page source: https://andromeda.ai/
They said they "believe that the total amount of computation required by ESM3 is the largest for any biological model ever created."
Inferring protein sequence, structure, and function
Language models that process text generally use tokens as basic units, but multimodal protein models are more complex and require converting sequences, three-dimensional structures, and functions into discrete letters for representation.
In order to better expand the training scale and unleash the "emergence" generation potential of the model, the vocabulary used by ESM3 can well connect sequences, structures and functions in the same language model for joint reasoning.
Different from language models such as GPT, the training objective of ESM3 inherits the masked language modeling objective.
Part of the sequence, structure, and function of each protein will be masked, and the model needs to gradually understand the deep connection between the three during training to predict the masked position. If the labels of all positions are masked, it is equivalent to performing a generation task.
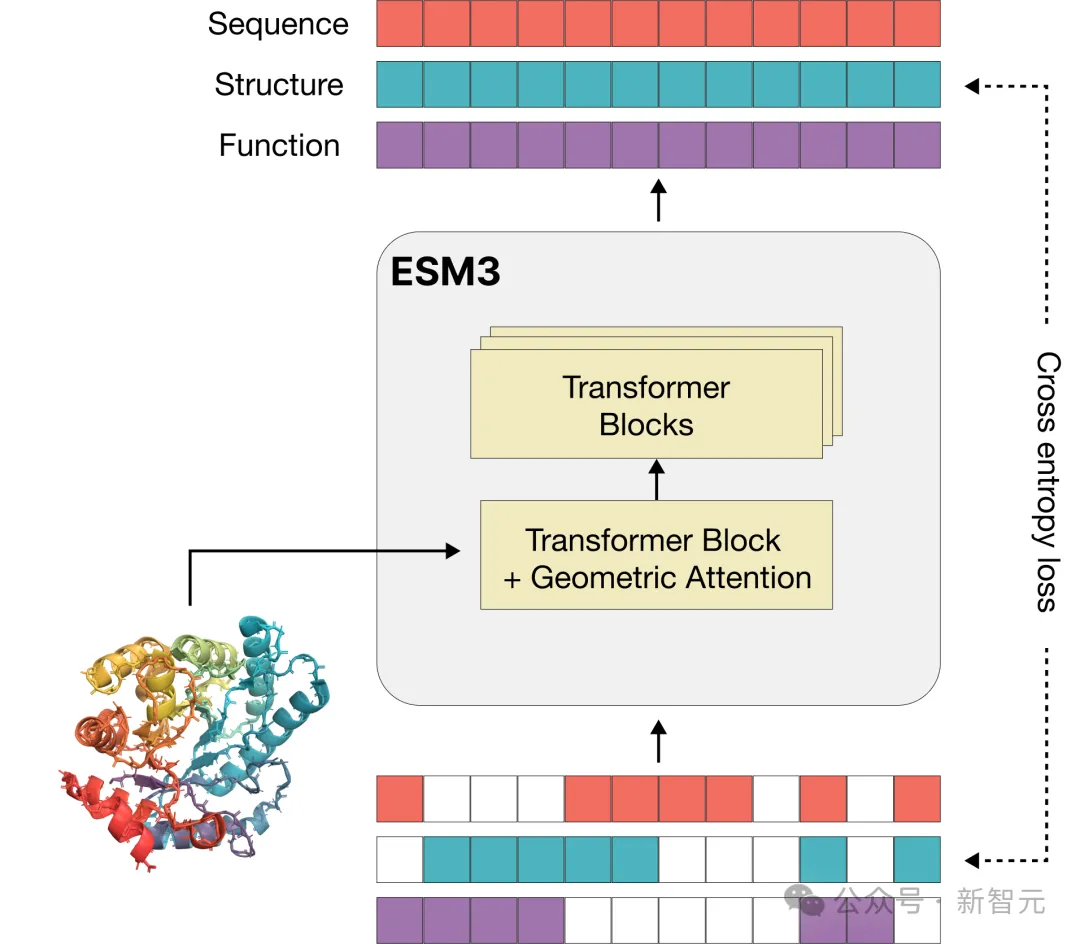
Since it is jointly trained on the sequence, structure, and function of proteins, these three modalities can be arbitrarily masked and predicted, so ESM3 achieves "all to all" prediction or generation.
That is, the model's input can be any combination of the three modes that are partially or fully specified. This powerful multimodal reasoning capability has great application value, allowing scientists to design completely new proteins with unprecedented flexibility and control.
For example, the model can be prompted to combine structure, sequence and function to propose a potential scaffold structure for the active site of PETase, a commonly used plastic. If PETase is successfully designed, it can be used to efficiently decompose plastic waste.

ESM3 scaffolds the active site of PETase through multimodal cues from sequence, structure, and function
Tom Sercu, co-founder and vice president of engineering at Evolutionary Scale, said that in internal testing, ESM showed impressive creativity in responding to a variety of complex prompts.
“It was able to solve an extremely difficult protein design problem and create a new type of green fluorescent protein. ESM3 can help scientists accelerate their work and open up new possibilities – we look forward to seeing its contribution to life science research in the future.”
The model also learns to simulate evolution when billions of proteins come from different positions on the evolutionary timeline, with rich diversity.
Capabilities emerge with scale
Just as LLM has "emerged" with capabilities such as language understanding and reasoning as it expands in scale, ESM3 has also gradually demonstrated its capabilities as its scale increases in solving challenging protein design tasks. One of the important capabilities is atomic-level coordination.
For example, the prompt might specify that two amino acids that make up a protein need to be close in sequence but far apart in the structure. This measures the model's ability to achieve atomic-level accuracy in the structure generation task.
This is critical for designing functional proteins, and ESM3's ability to solve such complex generation tasks can be gradually improved with increasing scale.
Moreover, after the training is completed, the capabilities of ESM3 can be further improved, and its mechanism is similar to the RLHF method commonly used by LLM.
But the difference is that ESM3 does not receive feedback from humans. Instead, it can evaluate its own generation quality and make self-improvements. It can also combine existing experimental data and wet experiment results to align ESM3's generation with biological results.
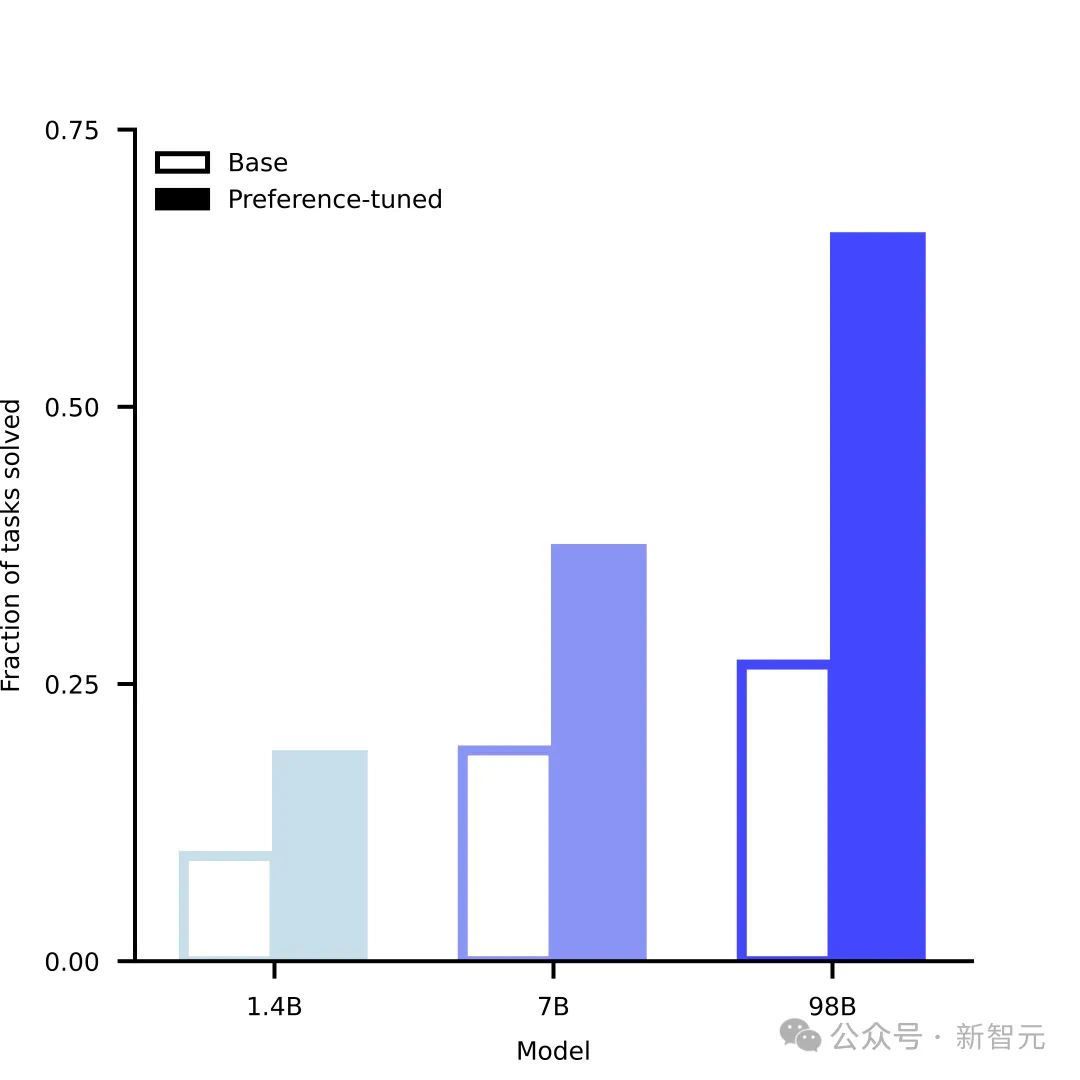
Model generation capabilities grow with scale, and fine-tuning improves performance significantly
Simulating 500 million years of evolution
In the published paper, the ESM3 team detailed the "simulated evolution" functions they observed in their model.

Paper address: https://evolutionaryscale-public.s3.us-east-2.amazonaws.com/research/esm3.pdf
Green Fluorescent Protein (GFP) and its family of fluorescent proteins are among the most beautiful proteins in nature, but they only exist in a few branches of the "Tree of Life".
But GFP is more than just beautiful. It contains a fluorescent chromophore, a molecule that absorbs short-wavelength monochromatic photons, captures some of the energy, and then releases another monochromatic photon with a longer wavelength. For example, naturally occurring GFP absorbs blue light and emits green light.
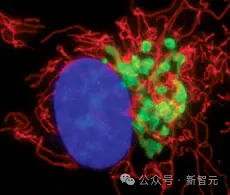
Due to this property, GFP can be used as a marker to help scientists observe proteins within cells, becoming one of the most widely used tools in biology. The discovery of GFP also won a Nobel Prize.
GFP is so unique and useful, and its structure is also very rare: a barrel-shaped structure composed of eleven chains with a helix running through the middle. After folding, the molecules in the center of the protein will rearrange themselves and produce a fluorescent chromophore.
This mechanism is unique. No other known protein can spontaneously form a fluorescent chromophore from its own structure, suggesting that even in nature, generating fluorescence is quite rare and difficult.
In order to have a wider range of applications in the laboratory, scientists have tried to add mutations or change colors and perform artificial synthesis. The latest machine learning technology can search for variants with sequence differences of up to 20%, but the main source of functional GFP is still nature rather than protein engineering.
It is not easy to find more variants in nature, because the evolution of new fluorescent proteins takes a long time - the family to which GFP belongs has a very long history, and the time when they diverged from the ancestral sequence can be traced back to hundreds of millions of years ago.
This thorny problem may be solved here in ESM3.
Taking the information of several sites in the natural GFP core structure as hints and using the CoT technique, ESM3 successfully generated candidates for novel GFPs.
Such a generation could not have been the result of random luck or global search, because the possible combinations of sequences and structures would reach an astronomical number - 20 229 x 4096 229 , more than the number of atoms in the visible universe combined.
In the first experiment, the team tested 96 candidate proteins generated by ESM3, among which a successful luminescent sample emerged with a very unique structure that was far from any protein found in nature.
In another set of 96 candidate samples, several proteins with brightness similar to natural GFP were found, among which the one with the highest brightness was named esmGFP, which had 96 mutations compared with the closest natural fluorescent protein (58% similarity in the sequence consisting of 229 amino acids).
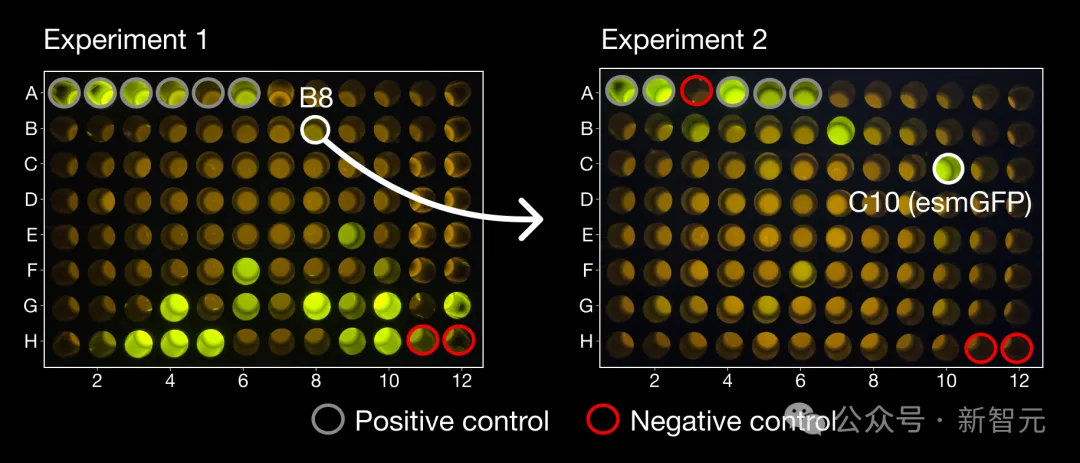
On the left is the product B8, which is very different from all known GFPs. Starting from B8, ESM3 produced the esmGFP on the right.
Unlike natural evolution, protein language models do not explicitly work within evolutionary constraints.
But in order for ESM3 to solve its training task of predicting the next mask token, the model must learn how evolution has evolved in the space of latent proteins.
In this sense, the process by which ESM3 generates esmGFP, which is very similar to the natural protein, can be viewed as an evolutionary simulator.
It is paradoxical to conduct traditional evolutionary analysis on esmGFP because it was created outside of natural processes, but it is still possible to gain insights from the tools of evolutionary biology into how long it takes for a protein to diverge from its closest sequence neighbors through natural evolution.
Therefore, the research team used evolutionary biology methods to analyze esmGFP as if it were a newly discovered protein in nature. They estimated that esmGFP is equivalent to more than 500 million years of natural evolution performed by the evolution simulator.

Rendering of esmGFP
Open Model
Since its inception, the ESM project has been committed to open science by publishing code and models. The code and model weights released by the team a few years ago can still be found on GitHub and HuggingFace.
Warehouse address: https://huggingface.co/facebook/esm2_t36_3B_UR50D/tree/main
It is amazing to see creative and impactful applications of the ESM model in research and industry:
- Hie et al. used ESM-1v and ESM-1b to evolve antibodies with improved therapeutically relevant properties such as binding affinity, thermal stability, and virus neutralization.
- BioNTech and InstaDeep fine-tuned an ESM language model for detecting variants in the COVID spike protein, successfully flagging all 16 variants of concern prior to WHO designation.
- Brandes et al. used ESM-1b to predict the clinical effect of mutations, which remains the strongest method for this important task.
- Marsiglia et al. used ESM-1v to engineer novel anti-CRISPR protein variants that reduced off-target side effects while maintaining on-target editing functionality.
- Shanker et al. used ESM-IF1 to guide the evolution of diverse proteins, including laboratory-validated antibodies that are highly effective against SARS-CoV-2.
- Yu et al. fine-tuned ESM-1b to predict the functions of enzymes, including rare and understudied enzymes, and validated the predictions experimentally.
- Rosen et al. used ESM2 embeddings to construct gene representations in single-cell grounded models.
- Høie et al. fine-tuned ESM-IF1 on antibody structures, achieving state-of-the-art performance in sequence recovery of CDR regions and designing antibodies with high binding affinity.
And these are just a small fraction of the amazing work built on the ESM platform!
Today, the team officially announced that it will release the weights and code of an ESM3 1.4B parameter version so that scientists and developers can build on the concepts and architecture of ESM3.