Beyond AF2? Iambic, NVIDIA, and Caltech develop multi-scale deep generative model for state-specific protein-ligand complex structure prediction

Binding complexes formed by proteins and small molecule ligands are ubiquitous and critical to life. Although scientists have recently made progress in protein structure prediction, existing algorithms cannot systematically predict binding ligand structures and their regulatory effects on protein folding.
To resolve this discrepancy, researchers from AI pharmaceutical company Iambic Therapeutics, Nvidia Corporation, and the California Institute of Technology proposed NeuralPLexer, a computational method that uses only protein sequences and ligand molecules Graph input directly predicts protein-ligand complex structures.
NeuralPLexer employs deep generative models to sample the three-dimensional structure of bound complexes and their conformational changes at atomic resolution. The model is based on a diffusion process that combines fundamental biophysical constraints and a multiscale geometric deep learning system to iteratively sample residue-level contact maps and all heavy atom coordinates in a hierarchical manner.
NeuralPLexer predictions are consistent with structure determination experiments of important targets in enzyme engineering and drug discovery, and have great potential to accelerate the design of functional proteins and small molecules at the proteome scale.
The study was titled "State-specific protein–ligand complex structure prediction with a multiscale deep generative model" and was published in "Nature Machine Intelligence" on February 12, 2024.

Static protein structure prediction is insufficient to support drug design
Deep learning has made tremendous progress in predicting protein structure from one-dimensional amino acid sequences. State-of-the-art protein structure prediction networks, such as AlphaFold2 (AF2), employ prediction pipelines based on evolutionary, physical, and geometric constraints on protein structures. Specifically, evolutionary constraints extracted from multiple sequence alignments (MSA) or protein language models (PLM) and specialized neural networks are systematically combined with sequence-based information and geometric representations to achieve end-to-end three-dimensional (3D) ) structure prediction.
Although highly successful in predicting protein static structures, this single structural formulation of the protein folding problem provides incomplete information about protein function and has been found to be insufficient for structure-based drug design.
Generative deep learning is an alternative paradigm
However, computational modeling of protein-ligand complexes that incorporate substantial changes in receptor conformation is hampered by the high cost of simulating slow protein state transitions. Recent developments in generative deep learning offer an alternative paradigm and have made substantial progress in understanding complex visual and language domains.
Two noteworthy strategies for generative modeling include (1) autoregressive models, widely employed in Transformer networks for sequence data (e.g., natural language and genomics), based on sequential processes; (2) diffusion-based generative models, utilizing A stochastic process generates data by sampling from a prior distribution and using a neural network to stepwise reverse the noisy process.
Scientists have demonstrated that deep generative models are capable of producing de novo designed proteins with experimentally validated functionality, including language models for protein sequence design and diffusion models for protein backbone generation. Diffusion models can effectively simulate molecular structures beyond the protein backbone, especially in molecular docking and structure-based drug design.
However, so far, no team has developed generative models that can directly predict binding complex structures at atomic resolution with an accuracy comparable to structure determination experiments.
Deep generative models predict protein-ligand complex structures
In the latest research, the Iambic, NVIDIA, and Caltech teams introduce NeuralPLexer, a computational system that uses deep generative models informed by biophysical inductive biases to predict protein-ligand complex structures. This method can directly generate a set of structures of binding complexes given a protein sequence and ligand molecular graph input, conditioned on auxiliary features obtained from PLM and template protein structures retrieved from experimentally resolved homologs or computational models.
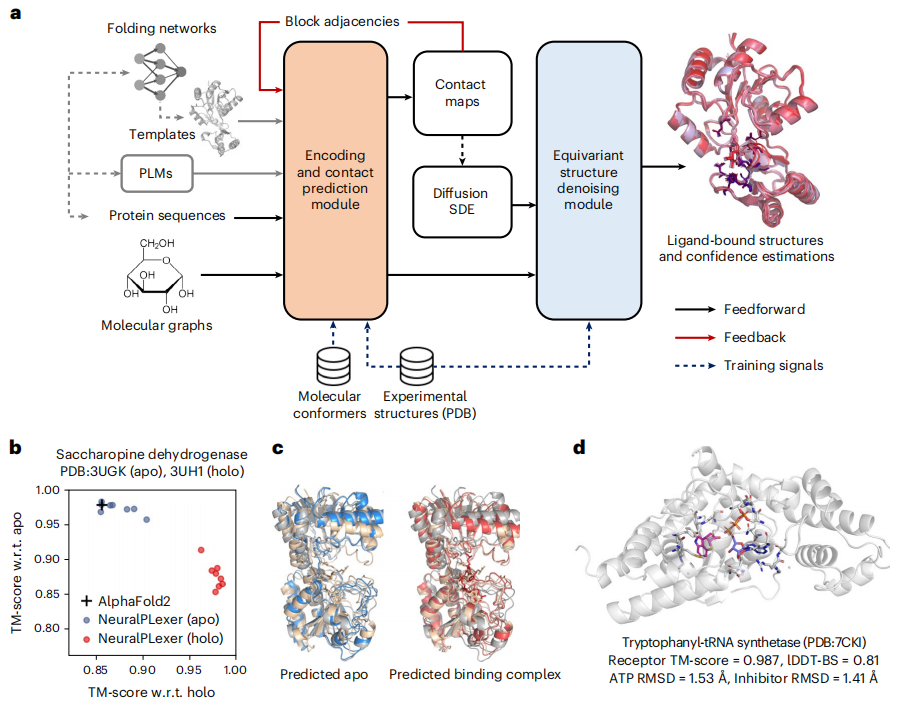
Illustration: NeuralPLexer accurately predicts structure and conformational changes in protein-ligand complexes. (Source: paper)
Both the prediction pipeline and the underlying neural network architecture are designed to reflect the multiscale hierarchical structure of biomolecular complexes. Specifically, NeuralPLexer includes:
(1) Graph-based networks that encode atomic-level chemical and geometric features of individual small molecule and amino acid graphs into tensor representations, implemented through a physics-inspired network architecture that is processed through millions of molecular conformation and biological activity databases training;
(2) Contact Prediction Module (CPM), motivated by recent visual language models and fold prediction networks, uses attention-based networks to generate residual-scale intermolecular distance distributions, coarse-grained contact maps, and associated pairwise representations;
(3) Equivariant Structure Denoising Module (ESDM) for generating combined complex atomic structures conditioned on the output of atomic-scale and residual-scale networks, using an equivariant structured denoising diffusion process and preserving proteins and ligands Chiral constraints on molecules.
When evaluated on blind protein-ligand docking, NeuralPLexer improved the prediction success rate by up to 78% compared to the best-performing existing method on the PDBBind2020 benchmark. In the design of ligand binding sites for challenging targets, NeuralPLexer can effectively recover up to 45% of the binding site structure using only computationally generated truncated scaffolds.
This represents a qualitative improvement in success rate compared to existing physics-based methods. Furthermore, NeuralPLexer demonstrates systematic advantages over existing methods in selectively predicting protein structures affected by induced fit binding or conformational selection; NeuralPLexer outperforms state-of-the-art methods on two benchmark datasets of ligand-binding proteins with large structural plasticity. The advanced protein structure prediction algorithm AF2, achieved the highest template modeling score (TM-score) (0.906 on average) and improved accuracy by 11-13% for domains that undergo significant conformational changes upon ligand binding.
NeuralPLexer's versatile ability to simulate ligand binding and protein structural changes enables rapid characterization of conformational landscapes, thereby facilitating a better understanding of the molecular mechanisms that control protein function, thereby aiding in the identification of unconventional approaches for therapeutic intervention and protein engineering at the proteome scale target.
Conclusion
As a data-driven approach, NeuralPLexer is versatile and can be continuously improved by integrating better experimental and bioinformatic data. Improvements in the curation of training and benchmarking datasets from the wider community may enable more systematic analysis of protein families for which there are no experimentally identified homologs and extend the method to more challenging systems such as post-translational modifications and polymorphic large heteromeric protein complexes.
This study provides a general computational framework for exploring these directions and paves the way for rapid and accurate protein-ligand complex structure prediction, thereby promoting advances in the fields of structural biology, drug discovery, and protein engineering.
Paper link: https://www.nature.com/articles/s42256-024-00792-z